Task #3: Design primers agagtctcct cagacgccga gatgctggtc atggcgcccc gaaccgtcct cctgctgctc 61 tcggcggccc tggccctgac cgagacctgg gccggtgagt gcgggtcggg agggaaatgg 121 cctctgccgg gaggagcgag gggaccgcag gcgggggcgc atgacctcag gagccgcgcc 181 gggaggaggg tcgggcgggt ctcagcccct cctcaccccc aggctcccac tccatgtggt 241 atttctacac ctccgtgtcc cggcccggcc gcggggagcc ccgcttcatc tcagtgggct 301 acgtggacga cacccagttc gtgaggttcg acagcgacgc cgcgagtccg agagaggagc 361 cgcgggcgcc gtggatagag caggaggggc cggagtattg ggaccggaac acacagatct 421 acaaggccca ggcacagact gaccgagaga gcctgcggaa cctgcgcttc tactacaacc 481 agagcgaggc cgttgcgtga ccccggcccg gggcgcaggt cacgactccc catcccccac 541 gtacggcccg ggtcgccccg agtctccggg tccgagatcc gcctccctga ggccgcggga a) First you'll need to design primers to PCR-amplify amino acids 45-56. i) Record the sequences of the forward and reverse primers, in the 5' to 3' direction. Each primer must be 8 nucleotides long (note that normally primers are much longer than this). Show intermediate steps in your primer design process and/or explain how you determined the primer sequences. ii) Using the method described in the pre-tutorial material, calculate the Tm for each primer (in °C). b) Next, you'll need to incorporate restriction sites into your primers. Shown here is the expression vector you want to clone the C-terminal domain into ("KanR2” is the antibiotic resistance gene): KanR2 pBR322 origin f1 origin T7 terminator Xhol (159) Eagl (167) NotI (167) HindIII (174) Sall (180) PET-28b(+) 5368 bp (Novagen) Sacl (191) EcoRI (193) Multiple Cloning Site (MCS) BamHI (199) Nhel (231) Ndel (238) Ncol (296) T7 promoter lacl i) Select a restriction enzyme (one that will generate sticky ends) that you could clone your fragment into, and indicate the sequence of its restriction site (e.g. in the format: Pstl, CTGCA/G). ii) Add the restriction enzyme site for the restriction enzyme you selected in (i) to each primer and re-write your primer sequences.
Task #3: Design primers agagtctcct cagacgccga gatgctggtc atggcgcccc gaaccgtcct cctgctgctc 61 tcggcggccc tggccctgac cgagacctgg gccggtgagt gcgggtcggg agggaaatgg 121 cctctgccgg gaggagcgag gggaccgcag gcgggggcgc atgacctcag gagccgcgcc 181 gggaggaggg tcgggcgggt ctcagcccct cctcaccccc aggctcccac tccatgtggt 241 atttctacac ctccgtgtcc cggcccggcc gcggggagcc ccgcttcatc tcagtgggct 301 acgtggacga cacccagttc gtgaggttcg acagcgacgc cgcgagtccg agagaggagc 361 cgcgggcgcc gtggatagag caggaggggc cggagtattg ggaccggaac acacagatct 421 acaaggccca ggcacagact gaccgagaga gcctgcggaa cctgcgcttc tactacaacc 481 agagcgaggc cgttgcgtga ccccggcccg gggcgcaggt cacgactccc catcccccac 541 gtacggcccg ggtcgccccg agtctccggg tccgagatcc gcctccctga ggccgcggga a) First you'll need to design primers to PCR-amplify amino acids 45-56. i) Record the sequences of the forward and reverse primers, in the 5' to 3' direction. Each primer must be 8 nucleotides long (note that normally primers are much longer than this). Show intermediate steps in your primer design process and/or explain how you determined the primer sequences. ii) Using the method described in the pre-tutorial material, calculate the Tm for each primer (in °C). b) Next, you'll need to incorporate restriction sites into your primers. Shown here is the expression vector you want to clone the C-terminal domain into ("KanR2” is the antibiotic resistance gene): KanR2 pBR322 origin f1 origin T7 terminator Xhol (159) Eagl (167) NotI (167) HindIII (174) Sall (180) PET-28b(+) 5368 bp (Novagen) Sacl (191) EcoRI (193) Multiple Cloning Site (MCS) BamHI (199) Nhel (231) Ndel (238) Ncol (296) T7 promoter lacl i) Select a restriction enzyme (one that will generate sticky ends) that you could clone your fragment into, and indicate the sequence of its restriction site (e.g. in the format: Pstl, CTGCA/G). ii) Add the restriction enzyme site for the restriction enzyme you selected in (i) to each primer and re-write your primer sequences.
Human Anatomy & Physiology (11th Edition)
11th Edition
ISBN:9780134580999
Author:Elaine N. Marieb, Katja N. Hoehn
Publisher:Elaine N. Marieb, Katja N. Hoehn
Chapter1: The Human Body: An Orientation
Section: Chapter Questions
Problem 1RQ: The correct sequence of levels forming the structural hierarchy is A. (a) organ, organ system,...
Related questions
Question
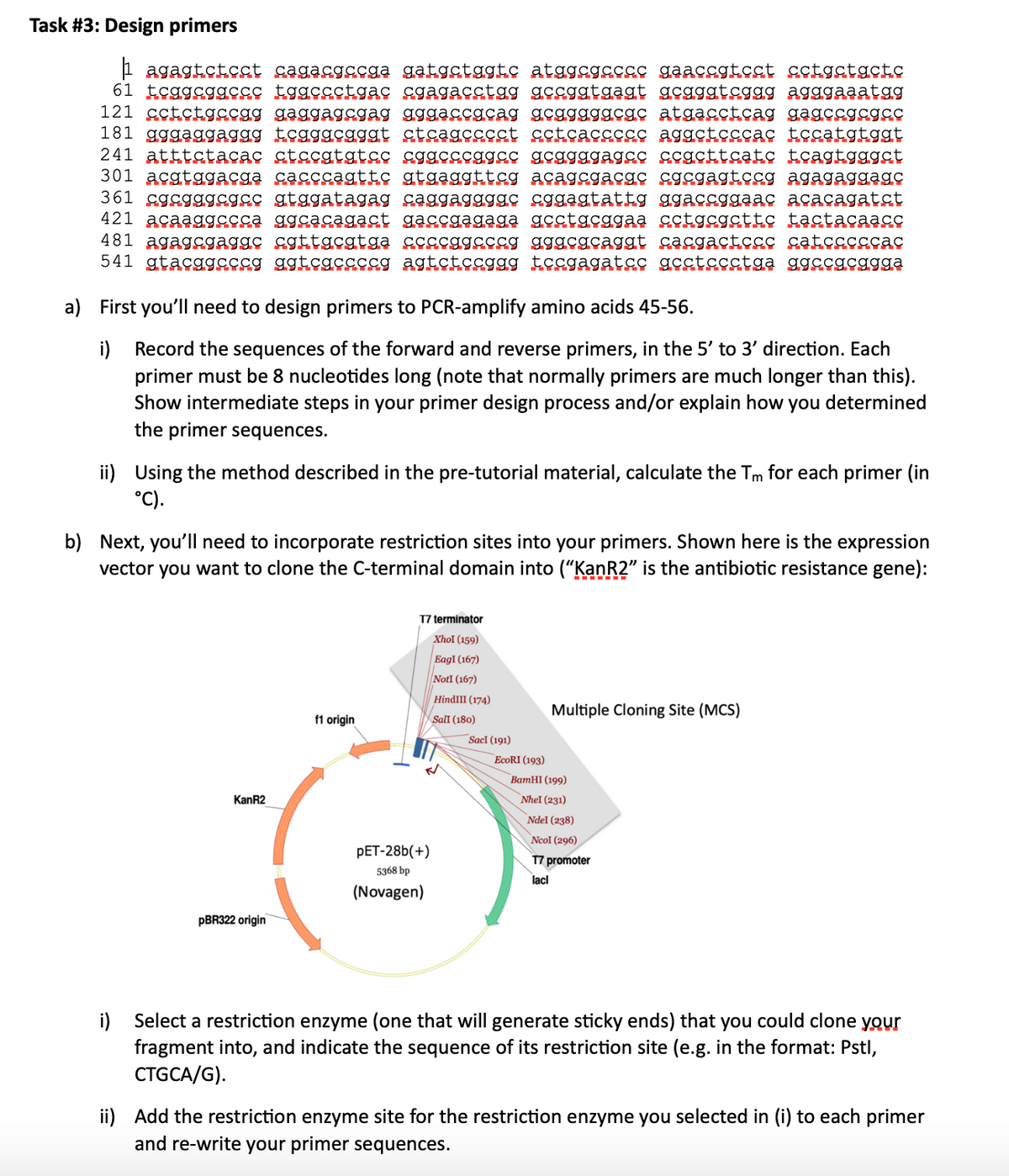
Transcribed Image Text:Task #3: Design primers
agagtctcct cagacgccga gatgctggtc atggcgcccc gaaccgtcct cctgctgctc
61 tcggcggccc tggccctgac cgagacctgg gccggtgagt gcgggtcggg agggaaatgg
121 cctctgccgg gaggagcgag gggaccgcag gcgggggcgc atgacctcag gagccgcgcc
181 gggaggaggg tcgggcgggt ctcagcccct cctcaccccc aggctcccac tccatgtggt
241 atttctacac ctccgtgtcc cggcccggcc gcggggagcc ccgcttcatc tcagtgggct
301 acgtggacga cacccagttc gtgaggttcg acagcgacgc cgcgagtccg agagaggagc
361 cgcgggcgcc gtggatagag caggaggggc cggagtattg ggaccggaac acacagatct
421 acaaggccca ggcacagact gaccgagaga gcctgcggaa cctgcgcttc tactacaacc
481 agagcgaggc cgttgcgtga ccccggcccg gggcgcaggt cacgactccc catcccccac
541 gtacggcccg ggtcgccccg agtctccggg tccgagatcc gcctccctga ggccgcggga
a) First you'll need to design primers to PCR-amplify amino acids 45-56.
i)
Record the sequences of the forward and reverse primers, in the 5' to 3' direction. Each
primer must be 8 nucleotides long (note that normally primers are much longer than this).
Show intermediate steps in your primer design process and/or explain how you determined
the primer sequences.
ii) Using the method described in the pre-tutorial material, calculate the Tm for each primer (in
°C).
b) Next, you'll need to incorporate restriction sites into your primers. Shown here is the expression
vector you want to clone the C-terminal domain into ("KanR2” is the antibiotic resistance gene):
KanR2
pBR322 origin
f1 origin
T7 terminator
Xhol (159)
Eagl (167)
NotI (167)
HindIII (174)
Sall (180)
PET-28b(+)
5368 bp
(Novagen)
Sacl (191)
EcoRI (193)
Multiple Cloning Site (MCS)
BamHI (199)
Nhel (231)
Ndel (238)
Ncol (296)
T7 promoter
lacl
i) Select a restriction enzyme (one that will generate sticky ends) that you could clone your
fragment into, and indicate the sequence of its restriction site (e.g. in the format: Pstl,
CTGCA/G).
ii) Add the restriction enzyme site for the restriction enzyme you selected in (i) to each primer
and re-write your primer sequences.
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 3 steps with 1 images

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Recommended textbooks for you

Human Anatomy & Physiology (11th Edition)
Biology
ISBN:
9780134580999
Author:
Elaine N. Marieb, Katja N. Hoehn
Publisher:
PEARSON

Biology 2e
Biology
ISBN:
9781947172517
Author:
Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:
OpenStax

Anatomy & Physiology
Biology
ISBN:
9781259398629
Author:
McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa Stouter
Publisher:
Mcgraw Hill Education,

Human Anatomy & Physiology (11th Edition)
Biology
ISBN:
9780134580999
Author:
Elaine N. Marieb, Katja N. Hoehn
Publisher:
PEARSON

Biology 2e
Biology
ISBN:
9781947172517
Author:
Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:
OpenStax

Anatomy & Physiology
Biology
ISBN:
9781259398629
Author:
McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa Stouter
Publisher:
Mcgraw Hill Education,

Molecular Biology of the Cell (Sixth Edition)
Biology
ISBN:
9780815344322
Author:
Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter Walter
Publisher:
W. W. Norton & Company

Laboratory Manual For Human Anatomy & Physiology
Biology
ISBN:
9781260159363
Author:
Martin, Terry R., Prentice-craver, Cynthia
Publisher:
McGraw-Hill Publishing Co.

Inquiry Into Life (16th Edition)
Biology
ISBN:
9781260231700
Author:
Sylvia S. Mader, Michael Windelspecht
Publisher:
McGraw Hill Education