Please complete questions (3), (4), and (5) in the screenshot provided into the code given in the other screenshot. Given below is the implementations of dijkstras and bellman ford already completed. Python 3.10 or later def bellman_ford(self,s) : """Bellman Ford Algorithm for single source shortest path. Keyword Arguments: s - The source vertex. """ distances = {v: float('inf') for v in self.adjacency_list} distances[s] = 0 parents = {v: None for v in self.adjacency_list} for _ in range(len(self.adjacency_list) - 1): for from_vertex in self.adjacency_list: for to_vertex in self.adjacency_list[from_vertex]: if distances[from_vertex] + self.weights[(from_vertex, to_vertex)] < distances[to_vertex]: distances[to_vertex] = distances[from_vertex] + self.weights[(from_vertex, to_vertex)] parents[to_vertex] = from_vertex for from_vertex in self.adjacency_list: for to_vertex in self.adjacency_list[from_vertex]: if distances[from_vertex] + self.weights[(from_vertex, to_vertex)] < distances[to_vertex]: # Negative cycle found, return empty list return [] # No negative cycles found, return list of 3-tuples return [(v, distances[v], parents[v]) for v in distances] def dijkstra(self,s) : """Dijkstra's Algorithm using a binary heap as the PQ. Keyword Arguments: s - The source vertex. """ distance = [math.inf for x in self._adj] parent = [None for x in self._adj] Q = PQ() distance[s] = 0 Q.add(s, 0) S = [] for u in range(len(self._adj)): if u != s: Q.add(u, math.inf) while not Q.is_empty(): u = Q.extract_min() S.append(u) for v, w in self._adj[u].__iter__(True): if (distance[u] + w) < distance[v]: parent[v] = u distance[v] = (distance[u] + w) returnlist = [] for v in S: returnlist.append((v, distance[v], parent[v])) return returnlist
Please complete questions (3), (4), and (5) in the screenshot provided into the code given in the other screenshot. Given below is the implementations of dijkstras and bellman ford already completed. Python 3.10 or later
def bellman_ford(self,s) :
"""Bellman Ford
Keyword Arguments:
s - The source vertex.
"""
distances = {v: float('inf') for v in self.adjacency_list}
distances[s] = 0
parents = {v: None for v in self.adjacency_list}
for _ in range(len(self.adjacency_list) - 1):
for from_vertex in self.adjacency_list:
for to_vertex in self.adjacency_list[from_vertex]:
if distances[from_vertex] + self.weights[(from_vertex, to_vertex)] < distances[to_vertex]:
distances[to_vertex] = distances[from_vertex] + self.weights[(from_vertex, to_vertex)]
parents[to_vertex] = from_vertex
for from_vertex in self.adjacency_list:
for to_vertex in self.adjacency_list[from_vertex]:
if distances[from_vertex] + self.weights[(from_vertex, to_vertex)] < distances[to_vertex]:
# Negative cycle found, return empty list
return []
# No negative cycles found, return list of 3-tuples
return [(v, distances[v], parents[v]) for v in distances]
def dijkstra(self,s) :
"""Dijkstra's Algorithm using a binary heap as the PQ.
Keyword Arguments:
s - The source vertex.
"""
distance = [math.inf for x in self._adj]
parent = [None for x in self._adj]
Q = PQ()
distance[s] = 0
Q.add(s, 0)
S = []
for u in range(len(self._adj)):
if u != s:
Q.add(u, math.inf)
while not Q.is_empty():
u = Q.extract_min()
S.append(u)
for v, w in self._adj[u].__iter__(True):
if (distance[u] + w) < distance[v]:
parent[v] = u
distance[v] = (distance[u] + w)
returnlist = []
for v in S:
returnlist.append((v, distance[v], parent[v]))
return returnlist
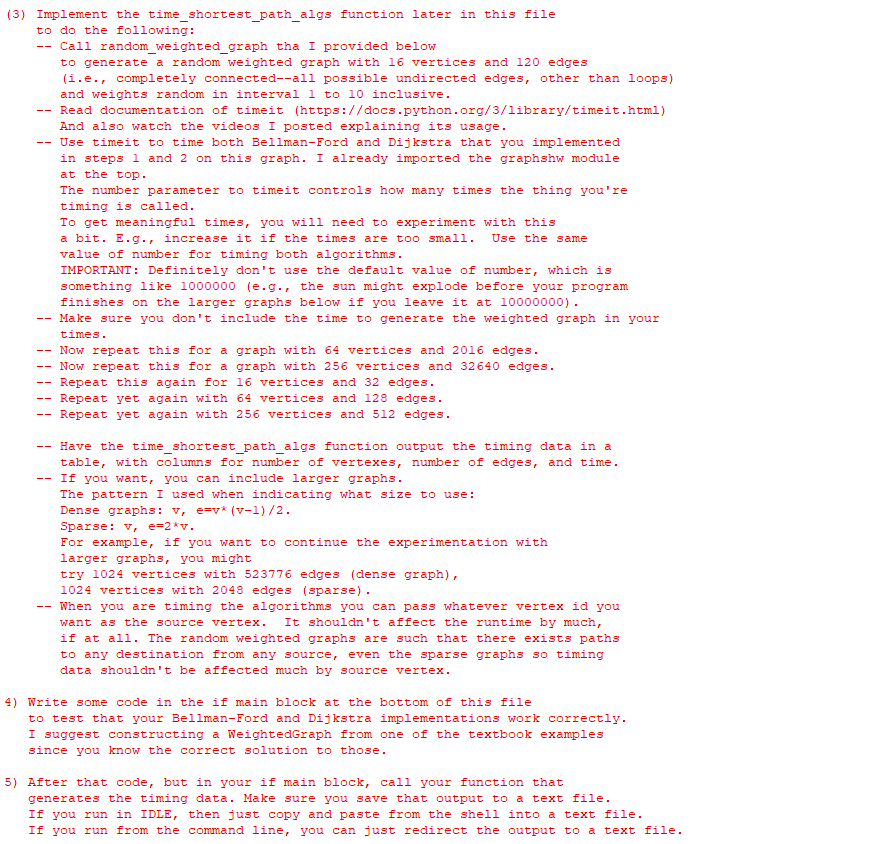
![def random_weighted_graph (v, e, min_w, max_w) :
"""Generates and returns a random weighted
graph with v vertices and e different edges.
Keyword arguments:
v number of vertices
e number of edges
if
min_w - minimum weight
max_w - maximum weight
edges = [ (random.randrange (0,1), i) for i in range (1, v) ]
# if desired number of edges greater than length of current edge list, then add more edges
if elen (edges) :
edgeSet = { x for x in edges }
not YetUsedEdges = [ (y,x) for x in range (1,v) for y in range (x) if (y,x) not in edgeSet]
random.shuffle (not YetUsedEdges)
count = e = len (edges)
count = min (count, len (not YetUsedEdges))
for i in range (count) :
#generate random edge weights
weights = [random.randint (min_w, max_w) for x in range (len (edges)) ]
edges.append (not YetUsedEdges.pop())
# construct a Digraph with the lists of edges and weights generated
G = WeightedGraph (v, edges, weights)
return G
def time_shortest_path_algs():
"""Generates a table of timing results comparing Dijkstra and Bellman-Ford."""
name
-
==
main":
#Here is where you write some code to test that your algorithms
#are correct.
#It is also where you will call your time_shortest_path_algs function.
#Don't forget to save output to a text file](/v2/_next/image?url=https%3A%2F%2Fcontent.bartleby.com%2Fqna-images%2Fquestion%2F0d59b49e-3987-421f-888e-fd2b8b6a0cc6%2F29f08746-4712-46e9-9a00-3505563b3898%2Frik8kf_processed.png&w=3840&q=75)
Step by step
Solved in 5 steps with 2 images


