Starting material: Tissue samples from four extinct species of moas were obtained from museum specimens. Tissue samples were also obtained from three species of kiwis, one emu, one cassowary, one ostrich, and two species of rhea. Experimental level Conceptual level 1. For cellular samples, treat with proteinase K (which digests protein) and a detergent that dissolves cell membranes. This releases the DNA Cells in Tissue tissue sample | Add proteinase K and detergent. from the cells. Isolate and purify the DNA released from the tissue. Chromosomal DNA Primers 2. Individually, mix the DNA samples with a pair of PCR primers that are complementary to the 12S FRNA gene. Add PCR primers. PCR DNA- PCR technique 3. Subject the samples to PCR. See Chapter 21 (Figure 21.5) for a description of PCR. Many copies of the 125 TRNA gene are made. 4. Subject the amplified DNA fragments to DNA sequencing. See Chapter 21 for a description of DNA sequencing. Sequence the amplified DNA. The amplification of the 12S IRNA gene allows it to be subjected to DNA sequencing. 5. Align the DNA sequences to each other. Methods of DNA sequence alienment are described in Chanter 24. Align sequences using computer programs. Align sequences to compare the degree of similarity.
Starting material: Tissue samples from four extinct species of moas were obtained from museum specimens. Tissue samples were also obtained from three species of kiwis, one emu, one cassowary, one ostrich, and two species of rhea. Experimental level Conceptual level 1. For cellular samples, treat with proteinase K (which digests protein) and a detergent that dissolves cell membranes. This releases the DNA Cells in Tissue tissue sample | Add proteinase K and detergent. from the cells. Isolate and purify the DNA released from the tissue. Chromosomal DNA Primers 2. Individually, mix the DNA samples with a pair of PCR primers that are complementary to the 12S FRNA gene. Add PCR primers. PCR DNA- PCR technique 3. Subject the samples to PCR. See Chapter 21 (Figure 21.5) for a description of PCR. Many copies of the 125 TRNA gene are made. 4. Subject the amplified DNA fragments to DNA sequencing. See Chapter 21 for a description of DNA sequencing. Sequence the amplified DNA. The amplification of the 12S IRNA gene allows it to be subjected to DNA sequencing. 5. Align the DNA sequences to each other. Methods of DNA sequence alienment are described in Chanter 24. Align sequences using computer programs. Align sequences to compare the degree of similarity.
Case Studies In Health Information Management
3rd Edition
ISBN:9781337676908
Author:SCHNERING
Publisher:SCHNERING
Chapter3: Informatics, Analytics, And Data use
Section: Chapter Questions
Problem 3.16.2C
Related questions
Question
From the results of the experiment , explain how we know that the kiwis are more closely related to the emu and cassowary than to the moas. Cite particular regions in the sequences that support your answer.
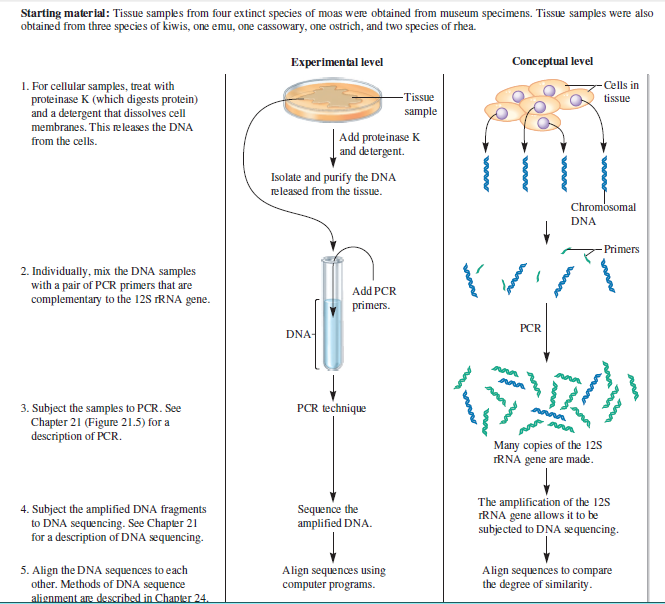
Transcribed Image Text:Starting material: Tissue samples from four extinct species of moas were obtained from museum specimens. Tissue samples were also
obtained from three species of kiwis, one emu, one cassowary, one ostrich, and two species of rhea.
Experimental level
Conceptual level
1. For cellular samples, treat with
proteinase K (which digests protein)
and a detergent that dissolves cell
membranes. This releases the DNA
Cells in
Tissue
tissue
sample
| Add proteinase K
and detergent.
from the cells.
Isolate and purify the DNA
released from the tissue.
Chromosomal
DNA
Primers
2. Individually, mix the DNA samples
with a pair of PCR primers that are
complementary to the 12S FRNA gene.
Add PCR
primers.
PCR
DNA-
PCR technique
3. Subject the samples to PCR. See
Chapter 21 (Figure 21.5) for a
description of PCR.
Many copies of the 125
TRNA gene are made.
4. Subject the amplified DNA fragments
to DNA sequencing. See Chapter 21
for a description of DNA sequencing.
Sequence the
amplified DNA.
The amplification of the 12S
IRNA gene allows it to be
subjected to DNA sequencing.
5. Align the DNA sequences to each
other. Methods of DNA sequence
alienment are described in Chanter 24.
Align sequences using
computer programs.
Align sequences to compare
the degree of similarity.
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by step
Solved in 2 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Recommended textbooks for you

Case Studies In Health Information Management
Biology
ISBN:
9781337676908
Author:
SCHNERING
Publisher:
Cengage

Case Studies In Health Information Management
Biology
ISBN:
9781337676908
Author:
SCHNERING
Publisher:
Cengage