10.18 Using the information in Figs. 10.14 and 10.18, explain how the two oligonucleotides 5'-CAAAGAAAAG-3' and 5'-CTTTTCTTTG-3' assemble into a double helical structure (see Fig. 10.14 for the 3' and 5' numbering, and definitions of C, A, G and T).
10.18 Using the information in Figs. 10.14 and 10.18, explain how the two oligonucleotides 5'-CAAAGAAAAG-3' and 5'-CTTTTCTTTG-3' assemble into a double helical structure (see Fig. 10.14 for the 3' and 5' numbering, and definitions of C, A, G and T).
Biology 2e
2nd Edition
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:Matthew Douglas, Jung Choi, Mary Ann Clark
Chapter15: Genes And Proteins
Section: Chapter Questions
Problem 22CTQ: If mRNA is complementary to the DNA template strand and the DNA template strand is complementary to...
Related questions
Question
![10.18 Using the information in Figs. 10.14 and
10.18, explain how the two oligonucleotides
5'-CAAAGAAAAG-3' and 5'-CTTTTCTTTG-3'
assemble into a double helical structure (see Fig.
10.14 for the 3' and 5' numbering, and definitions
of C, A, G and T).
Fig. 10.18. Two strands of oligonucleotides sequenced
5-CAAAGAAAAG-3' and 5'-CTTTTCTTTG-3' assemble into a
double helix. The structure has been determined by X-ray diffraction
[M. L. Kopka et al. (1996) J. Mol. Biol., vol. 334, p. 653]. The
backbone of each oligonucleotide is depicted as an arrow pointing
towards the C3' end of the sequence, and the nucleobases are shown in
a 'ladder' representation. The nucleobases are colour coded: G, green;
A, red; C, purple; T, turquoise.](/v2/_next/image?url=https%3A%2F%2Fcontent.bartleby.com%2Fqna-images%2Fquestion%2F00a3542e-c679-4030-a1b6-c1552b026fb7%2F367ba056-bfbe-47d7-b809-88573b31b37a%2Fexj38bb_processed.png&w=3840&q=75)
Transcribed Image Text:10.18 Using the information in Figs. 10.14 and
10.18, explain how the two oligonucleotides
5'-CAAAGAAAAG-3' and 5'-CTTTTCTTTG-3'
assemble into a double helical structure (see Fig.
10.14 for the 3' and 5' numbering, and definitions
of C, A, G and T).
Fig. 10.18. Two strands of oligonucleotides sequenced
5-CAAAGAAAAG-3' and 5'-CTTTTCTTTG-3' assemble into a
double helix. The structure has been determined by X-ray diffraction
[M. L. Kopka et al. (1996) J. Mol. Biol., vol. 334, p. 653]. The
backbone of each oligonucleotide is depicted as an arrow pointing
towards the C3' end of the sequence, and the nucleobases are shown in
a 'ladder' representation. The nucleobases are colour coded: G, green;
A, red; C, purple; T, turquoise.
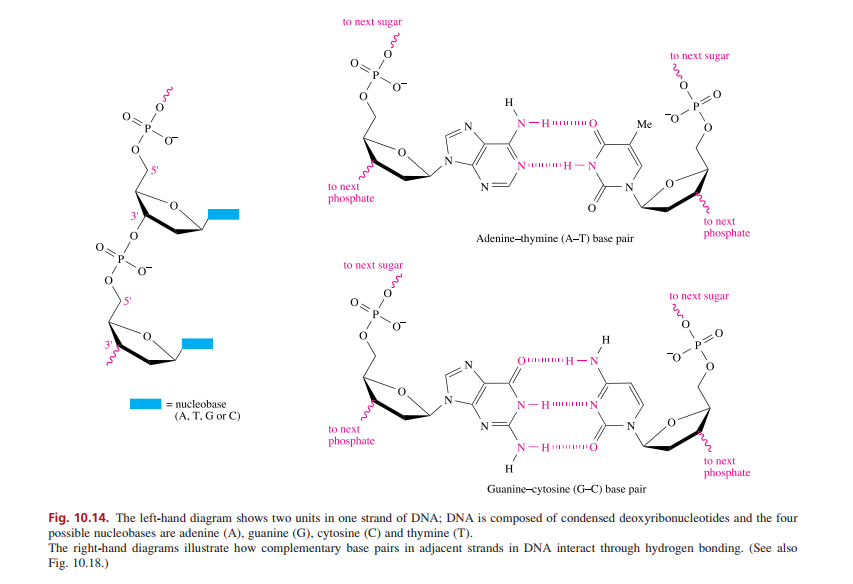
Transcribed Image Text:to next sugar
to next sugar
H.
p=0
N-H IIO
Me
5'
N H-N
to next
phosphate
to next
phosphate
Adenine-thymine (A-T) base pair
to next sugar
to next sugar
O.
H
O H-N
= nucleobase
N-H IN
(A, T, G or C)
to next
phosphate
N-H O
to next
H
phosphate
Guanine-cytosine (G-C) base pair
Fig. 10.14. The left-hand diagram shows two units in one strand of DNA; DNA is composed of condensed deoxyribonucleotides and the four
possible nucleobases are adenine (A), guanine (G), cytosine (C) and thymine (T).
The right-hand diagrams illustrate how complementary base pairs in adjacent strands in DNA interact through hydrogen bonding. (See also
Fig. 10.18.)
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 2 steps

Recommended textbooks for you

Biology 2e
Biology
ISBN:
9781947172517
Author:
Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:
OpenStax

Biology 2e
Biology
ISBN:
9781947172517
Author:
Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:
OpenStax