Case study 1 Melanoma A 50-year-old woman presented one year ago with a skin lesion, a biopsy was taken. and histological examination showed malignant melanoma. The patient had surgery to remove the lesion and appeared to be clear of the disease. The patient did not have targeted therapy at this stage. On year after surgery, she developed metastasis in the liver. Tumour biopsy confirmed melanoma and next generation sequencing analysis of the cancer genome was carried out. The results of the genomic analysis are included in the table below. The list of genomic variants produced using next generation sequencing. Chromosome Gene Function Zygosity ID COSV56056643 7934 COSV61684056 18921 COSV52662617 17p13 COSV104574314 16p13 BRAF SMAD4 TP53 AXIN1 Base change c.1799T>A missense Heterozygous missense Heterozygous c.1082G>A missense Heterozygous c.535C>T missense Heterozygous c.948G>A Amino acid change 1. Introduce the topic focusing on the molecular background of melanoma 2 p.V600E p.R361H p.H179Y p.G650S TASK You are required to analyse and interpret the genomic data linked to the case study and critically discuss how the genomic analysis would help in patient diagnosis and therapeutic management.
Case study 1 Melanoma A 50-year-old woman presented one year ago with a skin lesion, a biopsy was taken. and histological examination showed malignant melanoma. The patient had surgery to remove the lesion and appeared to be clear of the disease. The patient did not have targeted therapy at this stage. On year after surgery, she developed metastasis in the liver. Tumour biopsy confirmed melanoma and next generation sequencing analysis of the cancer genome was carried out. The results of the genomic analysis are included in the table below. The list of genomic variants produced using next generation sequencing. Chromosome Gene Function Zygosity ID COSV56056643 7934 COSV61684056 18921 COSV52662617 17p13 COSV104574314 16p13 BRAF SMAD4 TP53 AXIN1 Base change c.1799T>A missense Heterozygous missense Heterozygous c.1082G>A missense Heterozygous c.535C>T missense Heterozygous c.948G>A Amino acid change 1. Introduce the topic focusing on the molecular background of melanoma 2 p.V600E p.R361H p.H179Y p.G650S TASK You are required to analyse and interpret the genomic data linked to the case study and critically discuss how the genomic analysis would help in patient diagnosis and therapeutic management.
Surgical Tech For Surgical Tech Pos Care
5th Edition
ISBN:9781337648868
Author:Association
Publisher:Association
Chapter9: Surgical Pharmacology And Anesthesia
Section: Chapter Questions
Problem 9.5CS
Related questions
Question
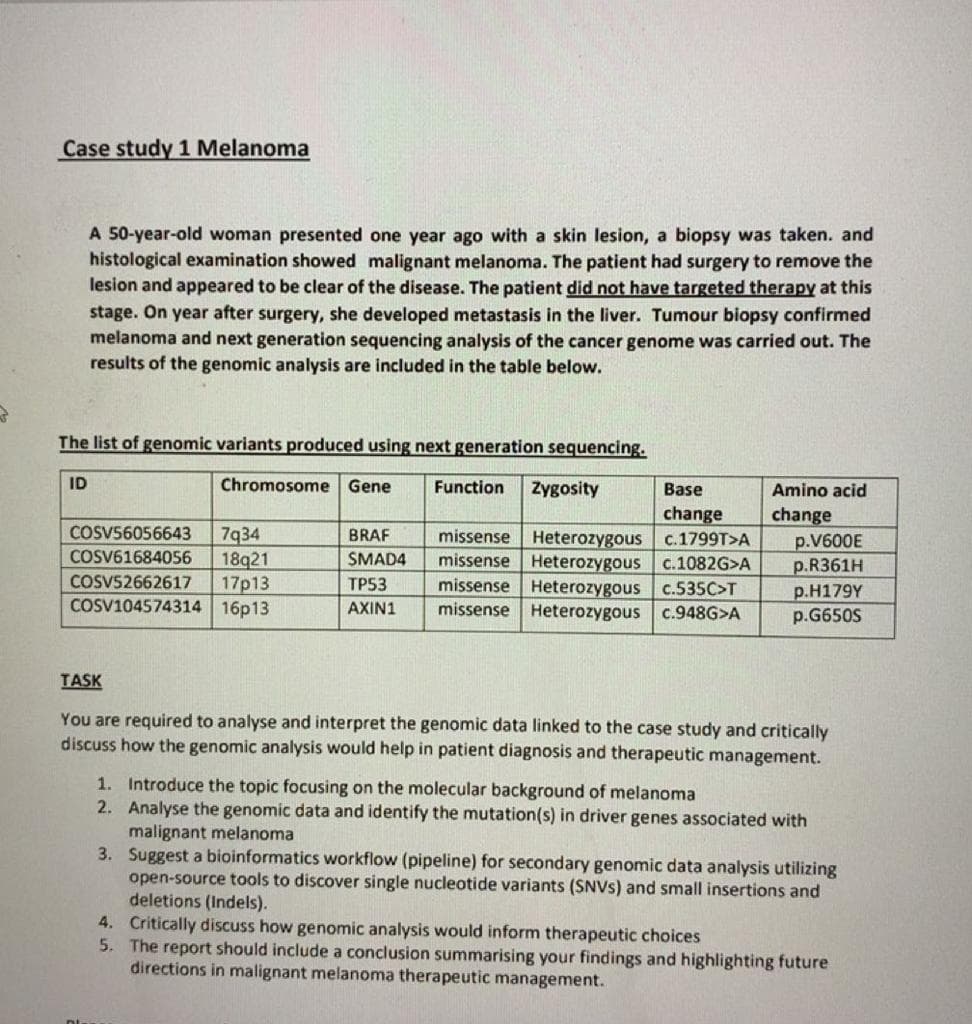
Transcribed Image Text:Case study 1 Melanoma
A 50-year-old woman presented one year ago with a skin lesion, a biopsy was taken. and
histological examination showed malignant melanoma. The patient had surgery to remove the
lesion and appeared to be clear of the disease. The patient did not have targeted therapy at this
stage. On year after surgery, she developed metastasis in the liver. Tumour biopsy confirmed
melanoma and next generation sequencing analysis of the cancer genome was carried out. The
results of the genomic analysis are included in the table below.
The list of genomic variants produced using next generation sequencing.
Chromosome Gene
Function Zygosity
ID
COSV56056643 7934
COSV61684056 18921
COSV52662617 17p13
COSV104574314 16p13
BRAF
SMAD4
TP53
AXIN1
Base
change
missense Heterozygous c.1799T>A
missense Heterozygous c.1082G>A
missense Heterozygous c.535C>T
missense Heterozygous c.948G>A
Amino acid
change
p.V600E
p.R361H
p.H179Y
p.G650S
TASK
You are required to analyse and interpret the genomic data linked to the case study and critically
discuss how the genomic analysis would help in patient diagnosis and therapeutic management.
1. Introduce the topic focusing on the molecular background of melanoma
2. Analyse the genomic data and identify the mutation(s) in driver genes associated with
malignant melanoma
3. Suggest a bioinformatics workflow (pipeline) for secondary genomic data analysis utilizing
open-source tools to discover single nucleotide variants (SNVS) and small insertions and
deletions (Indels).
4.
Critically discuss how genomic analysis would inform therapeutic choices
5. The report should include a conclusion summarising your findings and highlighting future
directions in malignant melanoma therapeutic management.
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 2 steps

Follow-up Questions
Read through expert solutions to related follow-up questions below.
Follow-up Question
I am confused about questions 2 and 3, especially 3, can someone help
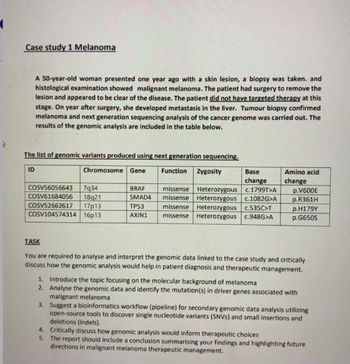
Transcribed Image Text:(
Case study 1 Melanoma
A 50-year-old woman presented one year ago with a skin lesion, a biopsy was taken. and
histological examination showed malignant melanoma. The patient had surgery to remove the
lesion and appeared to be clear of the disease. The patient did not have targeted therapy at this
stage. On year after surgery, she developed metastasis in the liver. Tumour biopsy confirmed
melanoma and next generation sequencing analysis of the cancer genome was carried out. The
results of the genomic analysis are included in the table below.
The list of genomic variants produced using next generation sequencing.
ID
Chromosome Gene
COSV56056643 7934
COSV61684056 18921
COSV52662617 17p13
COSV104574314 16p13
BRAF
SMAD4
TP53
AXIN1
Base
change
missense Heterozygous c.1799T>A
missense Heterozygous c.1082G>A
missense Heterozygous c.535C>T
missense Heterozygous c.948G>A
Function Zygosity
Amino acid
change
p.V600E
p.R361H
p.H179Y
p.G650S
TASK
You are required to analyse and interpret the genomic data linked to the case study and critically
discuss how the genomic analysis would help in patient diagnosis and therapeutic management.
1. Introduce the topic focusing on the molecular background of melanoma
2. Analyse the genomic data and identify the mutation(s) in driver genes associated with
malignant melanoma
3. Suggest a bioinformatics workflow (pipeline) for secondary genomic data analysis utilizing
open-source tools to discover single nucleotide variants (SNVS) and small insertions and
deletions (Indels).
4. Critically discuss how genomic analysis would inform therapeutic choices
5. The report should include a conclusion summarising your findings and highlighting future
directions in malignant melanoma therapeutic management.
Solution
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Recommended textbooks for you

Surgical Tech For Surgical Tech Pos Care
Health & Nutrition
ISBN:
9781337648868
Author:
Association
Publisher:
Cengage

Surgical Tech For Surgical Tech Pos Care
Health & Nutrition
ISBN:
9781337648868
Author:
Association
Publisher:
Cengage