7 8 9 9 10 11 1 TONSL vs Primary log2FPKM Raw counts 2 foldchange log2FC PValue KTB_103_pr KTB103_prir KTB103_prir KTB_103_TC KTB_103_TC KTB_103_TCKTB_103_pr KTB103_prir KTB103_prir KTB_103_TC KTB_103_TC KTB_103_TONSL-F_S22 3 3604.4176 11.81555 2.85E-284 1.55E-282-9.3129082 -9.3129082 -9.3129082 1.4648813 1.5160533 1.5272297 0315000 6 4 1099.1348 10.102153 4.99E-95 5.49E-94 -9.32157 -9.32157 -9.32157 -0.0733843 -0.3162863 -0.2790863 5 847.28604 9.7267053 1.49E-77 1.33E-76 -8.215888 -8.215888 -8.215888 0.631853 0.3966124 0.4940631 204 64000 6500306 memo 9.3744036 791.81993 9.6290286 8.59E-75 7.38E-74 -8.2711026 -8.2711026 -8.2711026 0.4262034 0.3831127 0.2707696 136.06836 EASDEES 726.06516 9.5039552 2.36E-69 1.87E-68 -6.8822306 -6.8822306 -6.8822306 1.5912286 1.7160924 1.5676387 600 4400302402006 4.36.363 ******* 2443603 **** 100*50*5.240*30 33103836 608.11897 9.2482098 1.2E-267 5.95E-266-7.9296227 -4.7142693 -7.9296227 2.9934394 3.2481759 3.2192825 www.c SERVEET MUO *600*65 0.0384336 0.000000 ***************** 456.80467 8.8354336 2.38E-43 -9.090888 -9.090888 -9.090888 -1.1241791 -1.4468333 -1.2171597 www.www 0:0934350 6104343 404247 076474 4064376* $0413500 C10423 384.39701 8.5864533 3.782E-39 1.792E-38 -9.5491347 -9.5491347 -9.5491347 -1.975471 -1.9613751 -1.9412599 www 367.82505 8.5228759 2.41E-37 11367.62505 6.5226159 ZNIES! 346.01397 8.4346865 1.09E-117 340.01997 0340009 4.56E-44 12 13 299.68622 8.2273089 4.24E-31 14 278.67084 8.1224182 3.13E-94 17 239.98475 7.9067989 4.70E-123 18 18 238.77865 1.10E-36 -8.6796094 -8.6796094 -8.6796094 -1.0796112 -1.2430199 -1.1466266 196094 -8.6796094 -1.0796112 -1.2430199 -1.1466266 BUCO HO 1.53E-116 -6.2563842 -8.8207137 -8.8207137 0.5240714 0.6412797 0.7507498 1.53E-110 -6.2563642 -6.8207137 -8.8207137 0.5240714 0.6412797 0.7507498 1.66E-30 -8.3760452 -8.3760452 -8.3760452 -1.1290682 -1.181848 -1.125163 -1.101646 -1.125163 3.41E-93-6.5188956 -6.5188956 -3.9634506 2.7864228 2.5182696 2.5766006 15 277.74252 8.1176043 1.11E-252 4.89E-251 -4.824966 -5.8989967 -5.7064293 2.602651 2.6864677 2.481924 16 10 259.38227 8.0189361 1.16E-26 4.07E-26 -8.4181912 -8.4181912 -8.4181912 -1.4660267 -1.5624062 -1.1988073 6.92E-122 -8.0754611 -4.8601077 -8.0754611 1.840504 1.6763788 1.4771684 -2.578528 1.86E-24 -6.424638 0.3993354 0.5234777 -6.424638 -6.424638 0475226 0.475236 6.0519258 1260692 04505629 02011205 2.06E-76 -8.6073708 -8.6073708 -6.0519258 0.1260692 0.4506639 0.2011305 3.26E-23 -9.1944906 -9.1944906 -9.1944906 -2.326509 -2.5992509 -2.21725 9.24E-133 -8.5795786 -8.5795786 -5.1523782 1.0913564 1.0728451 0.9667178 2.43E-22 -5.4410844 -5.4410844 -5.4410844 1.382889 1.035158 1.5361562 1.49E-21 -8.2515508 -8.2515508 -8.2515508 -1.1444625 -1.5463855 -1.8165229 2.19E-22 -9.8659117 -9.8659117 -9.8659117 -3.0419383 -3.0586068 -3.2266579 7.89953 19 236.26536 7.8842643 4485-24-9 5176038 -9 5176038 -9 5176038 -2.8844576 -2.4008619 20 330 12216 229.13316 7 8400424 1.36E-24 5.56E-25 7.8400424 2.31E-77 226.52461 7.823524 1.01E-23 220.17798 7.7825264 5.61E-134 219.35572 7.7771285 7.71E-23 21 22 22 23 24 217.6387 7.7657913 4.81E-22 25 215.16056 7.7492699 6.93E-23 DE_analysis + < H > K FDR R I 0 0 0 0 0 = 0 = 0 9 0 9 0 9 1 2 0 9 0 0 2 2 0 0 0 O 0 O 0 0 0 0 0 0 0 0 ◄ 0 0 0 0 0 = 2 • 0 9 0 9 0 9 0 9 0 0 0 1 + 0 2 0 0 O 0 0 0 0 0 O 0 0 0 0 0 0 0 = 0 0 = 0 9 0 9 0 9 0 9 0 0 1 ↓ 1 0 0 0 0 0 1 1 O 0 2 2 0 0 0 500 173 131 118 *** 101 553 71 54 55 185 43 180 531 35 275 28 32 121 33 232 32 39 32 475 134 102 105 101 *v* 605 52 50 45 43 184 38 137 516 30 225 36 32 139 25 210 23 27 29 557 160 127 113 106 690 71 2 59 34 56 30 231 46 166 521 45 228 37 36 36 136 38 227 38 26 30
Genetic Variation
Genetic variation refers to the variation in the genome sequences between individual organisms of a species. Individual differences or population differences can both be referred to as genetic variations. It is primarily caused by mutation, but other factors such as genetic drift and sexual reproduction also play a major role.
Quantitative Genetics
Quantitative genetics is the part of genetics that deals with the continuous trait, where the expression of various genes influences the phenotypes. Thus genes are expressed together to produce a trait with continuous variability. This is unlike the classical traits or qualitative traits, where each trait is controlled by the expression of a single or very few genes to produce a discontinuous variation.
the topic is Identification of Differentially Expressed Genes in Breast Cancer Using RNA-Seq
write a method section of a scientific report for this topic.
including the use of excel to get these result( will be included in the images) and also this website https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216238
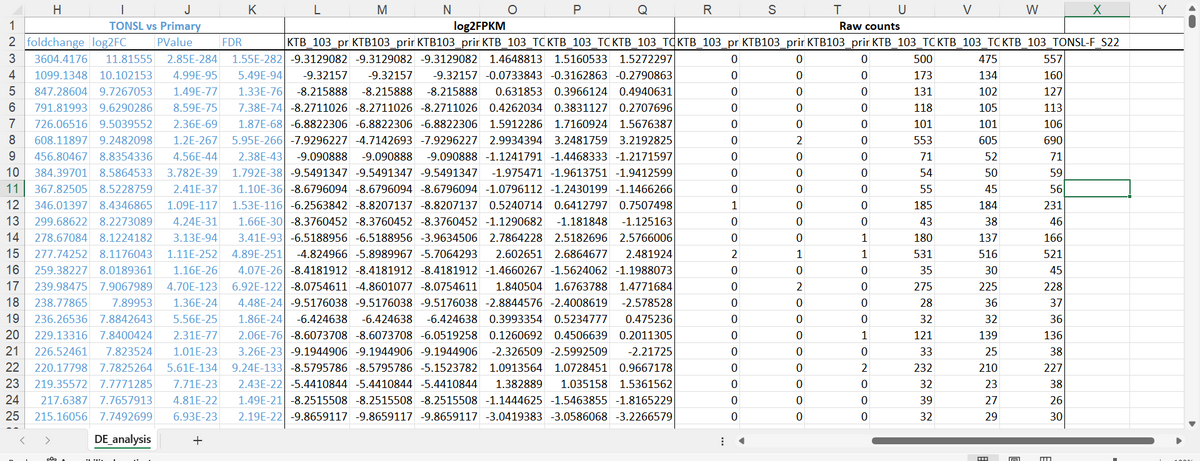
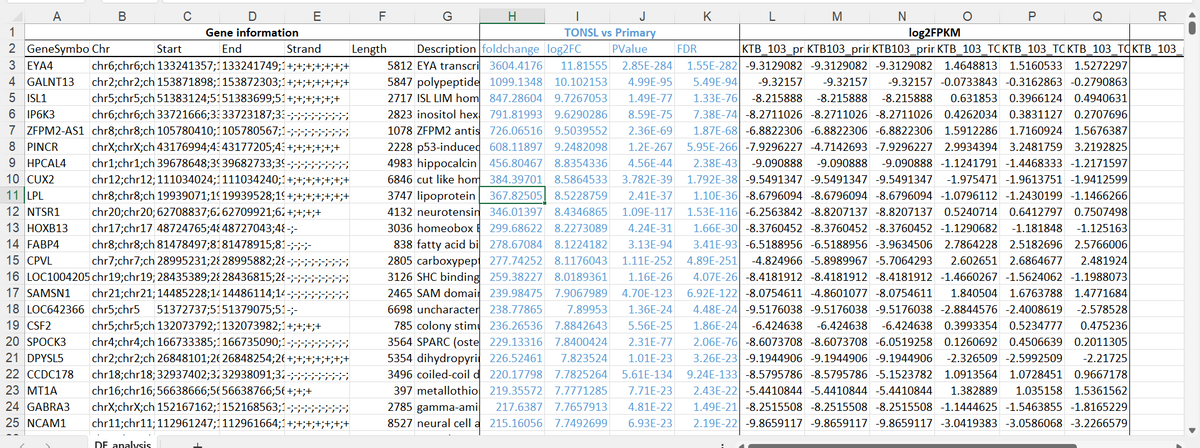
Step by step
Solved in 3 steps


