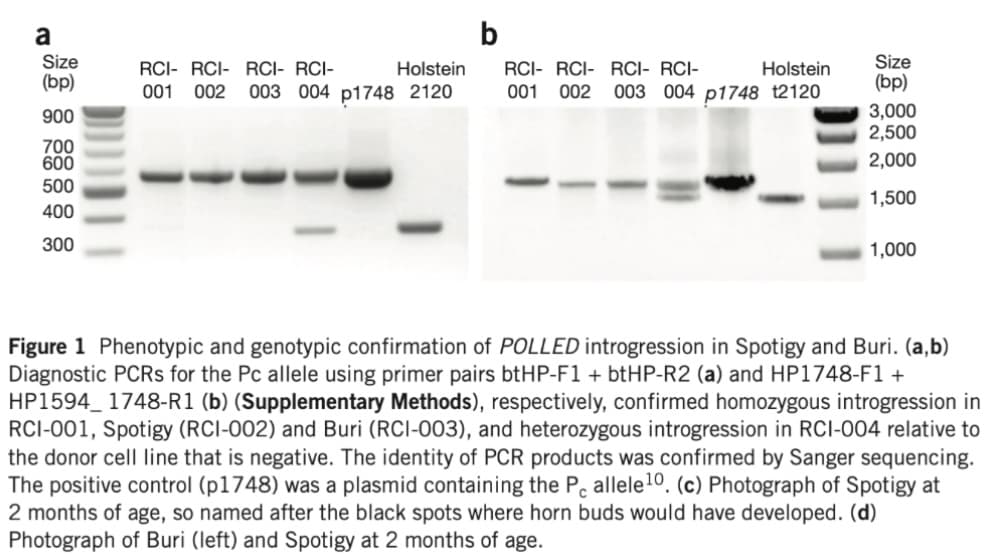
Using figure 1 and the following background information answer the following questions. Identification of the genetic cause of hornlessness in cattle has been the subject of intensive genetic and genomic research, culminating in the nomination of two different candidate neomutations on cattle chromosome 1 that are predicted to have arisen 500-1,000 years ago: a complex allele of Friesian origin (PF), an 80,128 base pair (bp) duplication (1909352–1989480 bp), and a second, simple allele of Celtic origin (PC) corresponding to a duplication of 212 bp (chromosome 1 positions 1705834–1706045) in place of a 10-bp deletion (1706051–1706060)We report the use of genome editing using transcription activator-like effector nucleases (TALENs) to introgress the putative PC POLLED allele into the genome of bovine embryo fibroblasts to try and produce a genotype identical to what is achievable using natural mating, but without the attendant genetic drag and admixture. In our previous studies, we used TALEN-stimulated homology-dependent repair to produce four cell lines either homozygous or heterozygous for the PC allele (Table 1). Each of the four lines were cloned by somatic cell nuclear transfer, and full-term pregnancies were established for three of the four lines. In total, five live calves were produced, representing two different dairy genetic backgrounds, 2122 and 2120 (Table 1). When calves were born, a board-certified veterinarian inspected each of the live-born calves visually and by palpation for horn buds, and observed none, which suggested polledness. Analysis of the polled genotype using diagnostic PCR1 confirmed the homozygous introgression of PC into RCI-001, RCI-002 (named Spotigy) and RCI-003 (named Buri), and heterozygous introgression into RCI-004 (RCI, Recombinetics, numbered in order of birth), whereas progenitor cells (2120) were negative for the PC allele (Fig. 1a,b). Two healthy, homozygous polled animals, Spotigy and Buri (Fig. 1c,d and Table 1), which are now more than 10 months old, were phenotypically polled. To evaluate off-target effects in two distinct edited lines, we sequenced the genomes of RCI-001 and RCI-002, derived from clones HP14-B4 and HP7-P4-A1 respectively, along with those of their progenitor cell lines, 2122 and 2120 respectively (Table 1). To avoid dehorning dairy cattle, transcription activator-like effector nucleases (TALENs) were used to edit the genome of bovine embryo fibroblasts with the Pc POLLED allele. After somatic cell nuclear transfer and full-term pregnancies, live calves were screened for the Pc POLLED allele. The results are shown in Figure 1. a. Results: Explain the data in terms of observations and patterns. Be sure to include information about all controls and relate results back to the predicted amplicon sizes. b. Conclusion: Describe how the data support or not support the hypothesis. c. Briefly describe a next experiment to be performed based on the conclusion.
Using figure 1 and the following background information answer the following questions. Identification of the genetic cause of hornlessness in cattle has been the subject of intensive genetic and genomic research, culminating in the nomination of two different candidate neomutations on cattle chromosome 1 that are predicted to have arisen 500-1,000 years ago: a complex allele of Friesian origin (PF), an 80,128 base pair (bp) duplication (1909352–1989480 bp), and a second, simple allele of Celtic origin (PC) corresponding to a duplication of 212 bp (chromosome 1 positions 1705834–1706045) in place of a 10-bp deletion (1706051–1706060)We report the use of genome editing using transcription activator-like effector nucleases (TALENs) to introgress the putative PC POLLED allele into the genome of bovine embryo fibroblasts to try and produce a genotype identical to what is achievable using natural mating, but without the attendant genetic drag and admixture. In our previous studies, we used TALEN-stimulated homology-dependent repair to produce four cell lines either homozygous or heterozygous for the PC allele (Table 1). Each of the four lines were cloned by somatic cell nuclear transfer, and full-term pregnancies were established for three of the four lines. In total, five live calves were produced, representing two different dairy genetic backgrounds, 2122 and 2120 (Table 1). When calves were born, a board-certified veterinarian inspected each of the live-born calves visually and by palpation for horn buds, and observed none, which suggested polledness. Analysis of the polled genotype using diagnostic PCR1 confirmed the homozygous introgression of PC into RCI-001, RCI-002 (named Spotigy) and RCI-003 (named Buri), and heterozygous introgression into RCI-004 (RCI, Recombinetics, numbered in order of birth), whereas progenitor cells (2120) were negative for the PC allele (Fig. 1a,b). Two healthy, homozygous polled animals, Spotigy and Buri (Fig. 1c,d and Table 1), which are now more than 10 months old, were phenotypically polled. To evaluate off-target effects in two distinct edited lines, we sequenced the genomes of RCI-001 and RCI-002, derived from clones HP14-B4 and HP7-P4-A1 respectively, along with those of their progenitor cell lines, 2122 and 2120 respectively (Table 1). To avoid dehorning dairy cattle, transcription activator-like effector nucleases (TALENs) were used to edit the genome of bovine embryo fibroblasts with the Pc POLLED allele. After somatic cell nuclear transfer and full-term pregnancies, live calves were screened for the Pc POLLED allele. The results are shown in Figure 1. a. Results: Explain the data in terms of observations and patterns. Be sure to include information about all controls and relate results back to the predicted amplicon sizes. b. Conclusion: Describe how the data support or not support the hypothesis. c. Briefly describe a next experiment to be performed based on the conclusion.
Human Heredity: Principles and Issues (MindTap Course List)
11th Edition
ISBN:9781305251052
Author:Michael Cummings
Publisher:Michael Cummings
Chapter10: From Proteins To Phenotypes
Section: Chapter Questions
Problem 3CS: A couple was referred for genetic counseling because they wanted to know the chances of having a...
Related questions
Topic Video
Question
100%
Using figure 1 and the following background information answer the following questions.
Identification of the genetic cause of hornlessness in cattle has been the subject of intensive genetic and genomic research, culminating in the nomination of two different candidate neomutations on cattle chromosome 1 that are predicted to have arisen 500-1,000 years ago: a complex allele of Friesian origin (PF), an 80,128 base pair (bp) duplication (1909352–1989480 bp), and a second, simple allele of Celtic origin (PC) corresponding to a duplication of 212 bp (chromosome 1 positions 1705834–1706045) in place of a 10-bp deletion (1706051–1706060)We report the use of genome editing using transcription activator-like effector nucleases (TALENs) to introgress the putative PC POLLED allele into the genome of bovine embryo fibroblasts to try and produce a genotype identical to what is achievable using natural mating, but without the attendant genetic drag and admixture. In our previous studies, we used TALEN-stimulated homology-dependent repair to produce four cell lines either homozygous or heterozygous for the PC allele (Table 1). Each of the four lines were cloned by somatic cell nuclear transfer, and full-term pregnancies were established for three of the four lines. In total, five live calves were produced, representing two different dairy genetic backgrounds, 2122 and 2120 (Table 1). When calves were born, a board-certified veterinarian inspected each of the live-born calves visually and by palpation for horn buds, and observed none, which suggested polledness. Analysis of the polled genotype using diagnostic PCR1 confirmed the homozygous introgression of PC into RCI-001, RCI-002 (named Spotigy) and RCI-003 (named Buri), and heterozygous introgression into RCI-004 (RCI, Recombinetics, numbered in order of birth), whereas progenitor cells (2120) were negative for the PC allele (Fig. 1a,b). Two healthy, homozygous polled animals, Spotigy and Buri (Fig. 1c,d and Table 1), which are now more than 10 months old, were phenotypically polled. To evaluate off-target effects in two distinct edited lines, we sequenced the genomes of RCI-001 and RCI-002, derived from clones HP14-B4 and HP7-P4-A1 respectively, along with those of their progenitor cell lines, 2122 and 2120 respectively (Table 1). To avoid dehorning dairy cattle, transcription activator-like effector nucleases (TALENs) were used to edit the genome of bovine embryo fibroblasts with the Pc POLLED allele. After somatic cell nuclear transfer and full-term pregnancies, live calves were screened for the Pc POLLED allele. The results are shown in Figure 1.
a. Results: Explain the data in terms of observations and patterns. Be sure to include information about all controls and relate results back to the predicted amplicon sizes.
b. Conclusion: Describe how the data support or not support the hypothesis.
c. Briefly describe a next experiment to be performed based on the conclusion.
Transcribed Image Text:2
b
Size
(bp)
RCI- RCI- RCI- RCI-
Holstein
RCI- RCI- RCI- RCI-
Size
Holstein
001 002 003 004 p1748 2120
001 002 003 004 p1748 12120
(bp)
900
3,000
2,500
700
600
2,000
500
1,500
400
300
1,000
Figure 1 Phenotypic and genotypic confirmation of POLLED introgression in Spotigy and Buri. (a,b)
Diagnostic PCRs for the Pc allele using primer pairs btHP-F1 + btHP-R2 (a) and HP1748-F1 +
HP1594 1748-R1 (b) (Supplementary Methods), respectively, confirmed homozygous introgression in
RCI-001, Spotigy (RCI-002) and Buri (RCI-003), and heterozygous introgression in RCI-004 relative to
the donor cell line that is negative. The identity of PCR products was confirmed by Sanger sequencing.
The positive control (p1748) was a plasmid containing the Pc allele 10. (c) Photograph of Spotigy at
2 months of age, so named after the black spots where horn buds would have developed. (d)
Photograph of Buri (left) and Spotigy at 2 months of age.
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 5 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Recommended textbooks for you

Human Heredity: Principles and Issues (MindTap Co…
Biology
ISBN:
9781305251052
Author:
Michael Cummings
Publisher:
Cengage Learning

Biology Today and Tomorrow without Physiology (Mi…
Biology
ISBN:
9781305117396
Author:
Cecie Starr, Christine Evers, Lisa Starr
Publisher:
Cengage Learning

Biology: The Unity and Diversity of Life (MindTap…
Biology
ISBN:
9781337408332
Author:
Cecie Starr, Ralph Taggart, Christine Evers, Lisa Starr
Publisher:
Cengage Learning

Human Heredity: Principles and Issues (MindTap Co…
Biology
ISBN:
9781305251052
Author:
Michael Cummings
Publisher:
Cengage Learning

Biology Today and Tomorrow without Physiology (Mi…
Biology
ISBN:
9781305117396
Author:
Cecie Starr, Christine Evers, Lisa Starr
Publisher:
Cengage Learning

Biology: The Unity and Diversity of Life (MindTap…
Biology
ISBN:
9781337408332
Author:
Cecie Starr, Ralph Taggart, Christine Evers, Lisa Starr
Publisher:
Cengage Learning