You are investigating the DNA-binding interaction of the drug candidate below, which binds to DNA via intercalation(" (H3C)2N N(CH3)2 a. You are lazy and don't want to do real computational simulations. You just estimate AHsinding based on the binding motif, shown below (drug is flat molecule in middle sandwiched between adjacent DNA base pairs). What would you estimate? Explain how you arrived at this number. (an aromatic interaction -5 kJ/mol)
You are investigating the DNA-binding interaction of the drug candidate below, which binds to DNA via intercalation(" (H3C)2N N(CH3)2 a. You are lazy and don't want to do real computational simulations. You just estimate AHsinding based on the binding motif, shown below (drug is flat molecule in middle sandwiched between adjacent DNA base pairs). What would you estimate? Explain how you arrived at this number. (an aromatic interaction -5 kJ/mol)
Biology: The Dynamic Science (MindTap Course List)
4th Edition
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Chapter19: Genomes And Proteomes
Section: Chapter Questions
Problem 1TYK
Related questions
Question
6
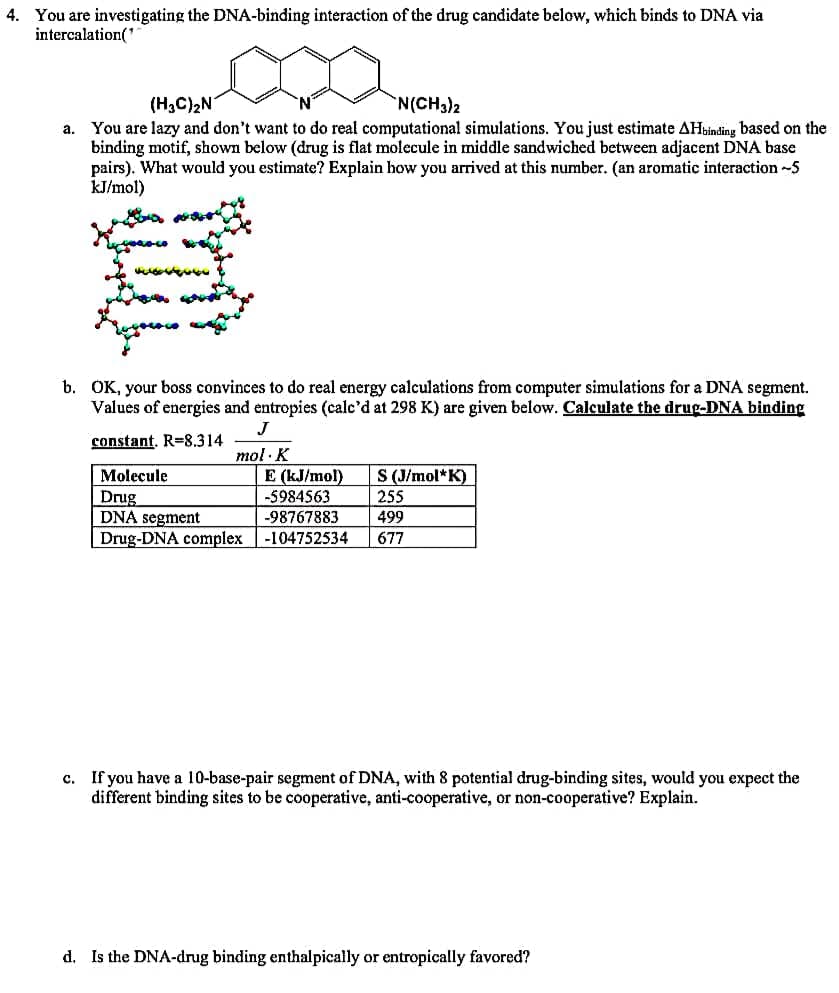
Transcribed Image Text:4. You are investigating the DNA-binding interaction of the drug candidate below, which binds to DNA via
intercalation('
(H3C)2N
You are lazy and don't want to do real computational simulations. You just estimate AHbinding based on the
binding motif, shown below (drug is flat molecule in middle sandwiched between adjacent DNA base
pairs). What would you estimate? Explain how you arrived at this number. (an aromatic interaction -5
kJ/mol)
N(CH3)2
а.
b. OK, your boss convinces to do real energy calculations from computer simulations for a DNA segment.
Values of energies and entropies (calc'd at 298 K) are given below. Calculate the drug-DNA binding
J
constant. R=8.314
mol · K
E (kJ/mol)
-5984563
Molecule
S (J/mol*K)
Drug
DNA segment
Drug-DNA complex-104752534
255
-98767883
499
677
c. If you have a 10-base-pair segment of DNA, with 8 potential drug-binding sites, would you expect the
different binding sites to be cooperative, anti-cooperative, or non-cooperative? Explain.
d. Is the DNA-drug binding enthalpically or entropically favored?
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 3 steps

Recommended textbooks for you

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:
9781305389892
Author:
Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:
Cengage Learning

Biology (MindTap Course List)
Biology
ISBN:
9781337392938
Author:
Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. Berg
Publisher:
Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:
9781305389892
Author:
Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:
Cengage Learning

Biology (MindTap Course List)
Biology
ISBN:
9781337392938
Author:
Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. Berg
Publisher:
Cengage Learning