Correct answers already provided! Please don't just tell me the answers bc I know them already. Help me with my own question. I get everything else in this problem other than the third option: Introduce the mutant human HD allele as a transgene into the mouse genome with transgene integration anywhere in the mouse genome. Why is the first question (left) okay with introducing mutant human HD allele and the second question (right) is not? I heard that introducing allele without using CRISPR-Cas9 is very rare and difficult. If so, how does it work in the first problem (Hungtinton's chorea)?
Correct answers already provided! Please don't just tell me the answers bc I know them already. Help me with my own question. I get everything else in this problem other than the third option: Introduce the mutant human HD allele as a transgene into the mouse genome with transgene integration anywhere in the mouse genome. Why is the first question (left) okay with introducing mutant human HD allele and the second question (right) is not? I heard that introducing allele without using CRISPR-Cas9 is very rare and difficult. If so, how does it work in the first problem (Hungtinton's chorea)?
Human Heredity: Principles and Issues (MindTap Course List)
11th Edition
ISBN:9781305251052
Author:Michael Cummings
Publisher:Michael Cummings
Chapter13: An Introduction To Genetic Technology
Section: Chapter Questions
Problem 19QP
Related questions
Question
Correct answers already provided! Please don't just tell me the answers bc I know them already. Help me with my own question.
I get everything else in this problem other than the third option: Introduce the mutant human HD allele as a transgene into the mouse genome with transgene integration anywhere in the mouse genome.
Why is the first question (left) okay with introducing mutant human HD allele and the second question (right) is not? I heard that introducing allele without using CRISPR-Cas9 is very rare and difficult. If so, how does it work in the first problem (Hungtinton's chorea)?
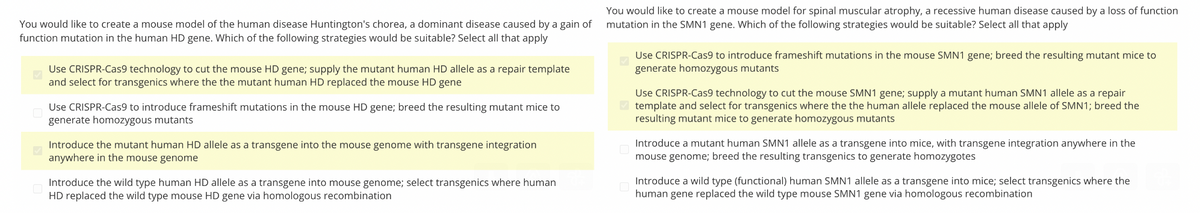
Transcribed Image Text:You would like to create a mouse model for spinal muscular atrophy, a recessive human disease caused by a loss of function
You would like to create a mouse model of the human disease Huntington's chorea, a dominant disease caused by a gain of mutation in the SMN1 gene. Which of the following strategies would be suitable? Select all that apply
function mutation in the human HD gene. Which of the following strategies would be suitable? Select all that apply
Use CRISPR-Cas9 technology to cut the mouse HD gene; supply the mutant human HD allele as a repair template
and select for transgenics where the the mutant human HD replaced the mouse HD gene
Use CRISPR-Cas9 to introduce frameshift mutations in the mouse HD gene; breed the resulting mutant mice to
generate homozygous mutants
Introduce the mutant human HD allele as a transgene into the mouse genome with transgene integration
anywhere in the mouse genome
Introduce the wild type human HD allele as a transgene into mouse genome; select transgenics where human
HD replaced the wild type mouse HD gene via homologous recombination
Use CRISPR-Cas9 to introduce frameshift mutations in the mouse SMN1 gene; breed the resulting mutant mice to
generate homozygous mutants
Use CRISPR-Cas9 technology to cut the mouse SMN1 gene; supply a mutant human SMN1 allele as a repair
template and select for transgenics where the the human allele replaced the mouse allele of SMN1; breed the
resulting mutant mice to generate homozygous mutants
Introduce a mutant human SMN1 allele as a transgene into mice, with transgene integration anywhere in the
mouse genome; breed the resulting transgenics to generate homozygotes
Introduce a wild type (functional) human SMN1 allele as a transgene into mice; select transgenics where the
human gene replaced the wild type mouse SMN1 gene via homologous recombination
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by step
Solved in 3 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Recommended textbooks for you

Human Heredity: Principles and Issues (MindTap Co…
Biology
ISBN:
9781305251052
Author:
Michael Cummings
Publisher:
Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:
9781305389892
Author:
Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:
Cengage Learning

Human Heredity: Principles and Issues (MindTap Co…
Biology
ISBN:
9781305251052
Author:
Michael Cummings
Publisher:
Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:
9781305389892
Author:
Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:
Cengage Learning