D. How many times would you expect to find a specific 20 base pair sequence in the human genome?
D. How many times would you expect to find a specific 20 base pair sequence in the human genome?
Human Heredity: Principles and Issues (MindTap Course List)
11th Edition
ISBN:9781305251052
Author:Michael Cummings
Publisher:Michael Cummings
Chapter13: An Introduction To Genetic Technology
Section: Chapter Questions
Problem 19QP
Related questions
Question

Transcribed Image Text:Student Guide
D. How many times would you expect to find a specific 20 base pair sequence in the
human genome?
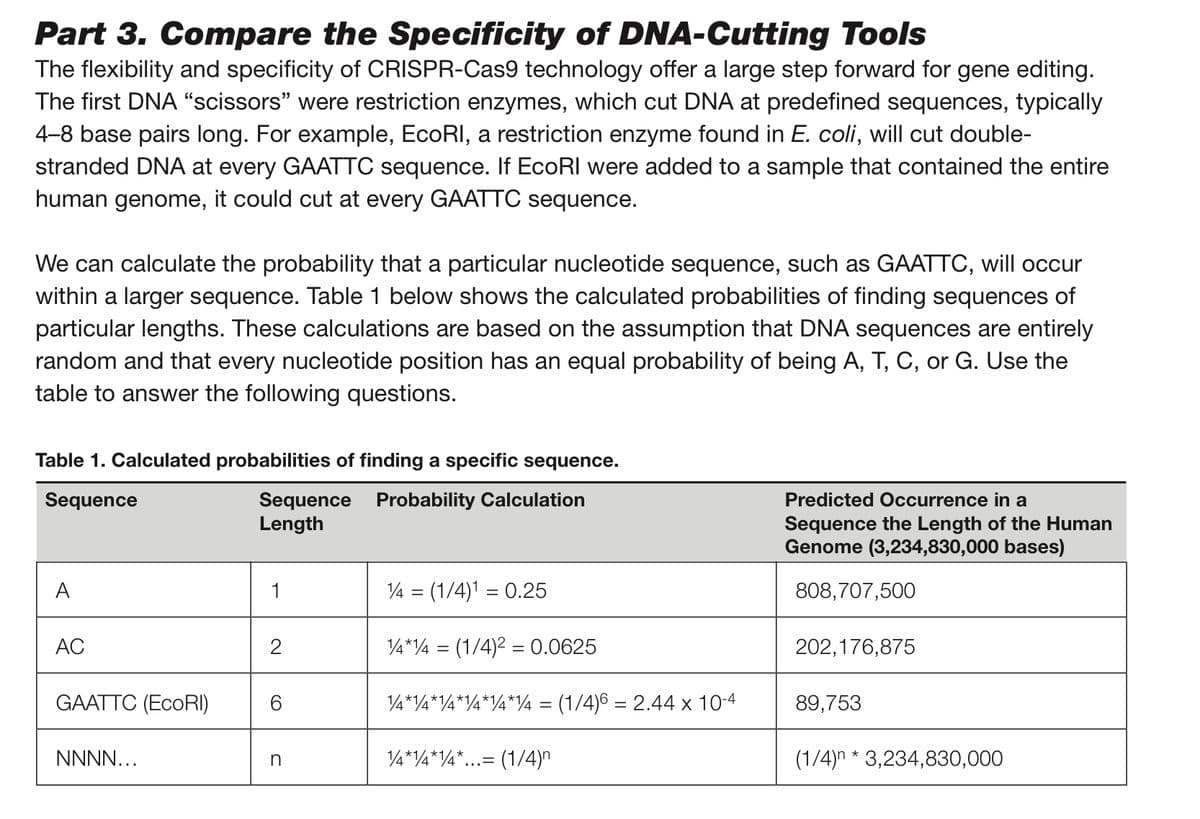
Transcribed Image Text:Part 3. Compare the Specificity of DNA-Cutting Tools
The flexibility and specificity of CRISPR-Cas9 technology offer a large step forward for gene editing.
The first DNA "scissors" were restriction enzymes, which cut DNA at predefined sequences, typically
4-8 base pairs long. For example, EcoRI, a restriction enzyme found in E. coli, will cut double-
stranded DNA at every GAATTC sequence. If EcoRI were added to a sample that contained the entire
human genome, it could cut at every GAATTC sequence.
We can calculate the probability that a particular nucleotide sequence, such as GAATTC, will occur
within a larger sequence. Table 1 below shows the calculated probabilities of finding sequences of
particular lengths. These calculations are based on the assumption that DNA sequences are entirely
random and that every nucleotide position has an equal probability of being A, T, C, or G. Use the
table to answer the following questions.
Table 1. Calculated probabilities of finding a specific sequence.
Sequence
Sequence Probability Calculation
Length
A
AC
GAATTC (EcoRI)
NNNN...
1
2
CO
n
14 = (1/4)¹ = 0.25
1/4*4 = (1/4)² = 0.0625
¼*14*¼*¼*¼*¼ = (1/4)6 = 2.44 x 10-4
14*14*14*. (1/4)
Predicted Occurrence in a
Sequence the Length of the Human
Genome (3,234,830,000 bases)
808,707,500
202,176,875
89,753
(1/4)n* 3,234,830,000
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 3 steps

Recommended textbooks for you

Human Heredity: Principles and Issues (MindTap Co…
Biology
ISBN:
9781305251052
Author:
Michael Cummings
Publisher:
Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:
9781305389892
Author:
Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:
Cengage Learning

Human Heredity: Principles and Issues (MindTap Co…
Biology
ISBN:
9781305251052
Author:
Michael Cummings
Publisher:
Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:
9781305389892
Author:
Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:
Cengage Learning