Figure 6-33 (Subunit interactions in an allosteric enzyme, and interactions with inhibitors and activators) depicts allosteric modulation of an enzyme through non-covalent interactions. What features of an activator might lead to different levels of enzyme regulation? Select one or more: a. ability to cause a conformational change that results in an altered activity. b. affinity for the regulatory site c. boiling point d. bond flexibility (i.e. abundance of freely rotating single bonds instead of more rigid bonds like double bonds) e. molecular weight
Figure 6-33 (Subunit interactions in an allosteric enzyme, and interactions with inhibitors and activators) depicts allosteric modulation of an enzyme through non-covalent interactions. What features of an activator might lead to different levels of enzyme regulation? Select one or more: a. ability to cause a conformational change that results in an altered activity. b. affinity for the regulatory site c. boiling point d. bond flexibility (i.e. abundance of freely rotating single bonds instead of more rigid bonds like double bonds) e. molecular weight
Human Physiology: From Cells to Systems (MindTap Course List)
9th Edition
ISBN:9781285866932
Author:Lauralee Sherwood
Publisher:Lauralee Sherwood
Chapter16: The Digestive System
Section: Chapter Questions
Problem 4TAHL
Related questions
Question
Figure 6-33 (Subunit interactions in an allosteric enzyme, and interactions with inhibitors and activators) depicts allosteric modulation of an enzyme through non-covalent interactions. What features of an activator might lead to different levels of enzyme regulation?
Select one or more:
a.
ability to cause a conformational change that results in an altered activity.
b.
affinity for the regulatory site
c.
boiling point
d.
bond flexibility (i.e. abundance of freely rotating single bonds instead of more rigid bonds like double bonds)
e.
molecular weight
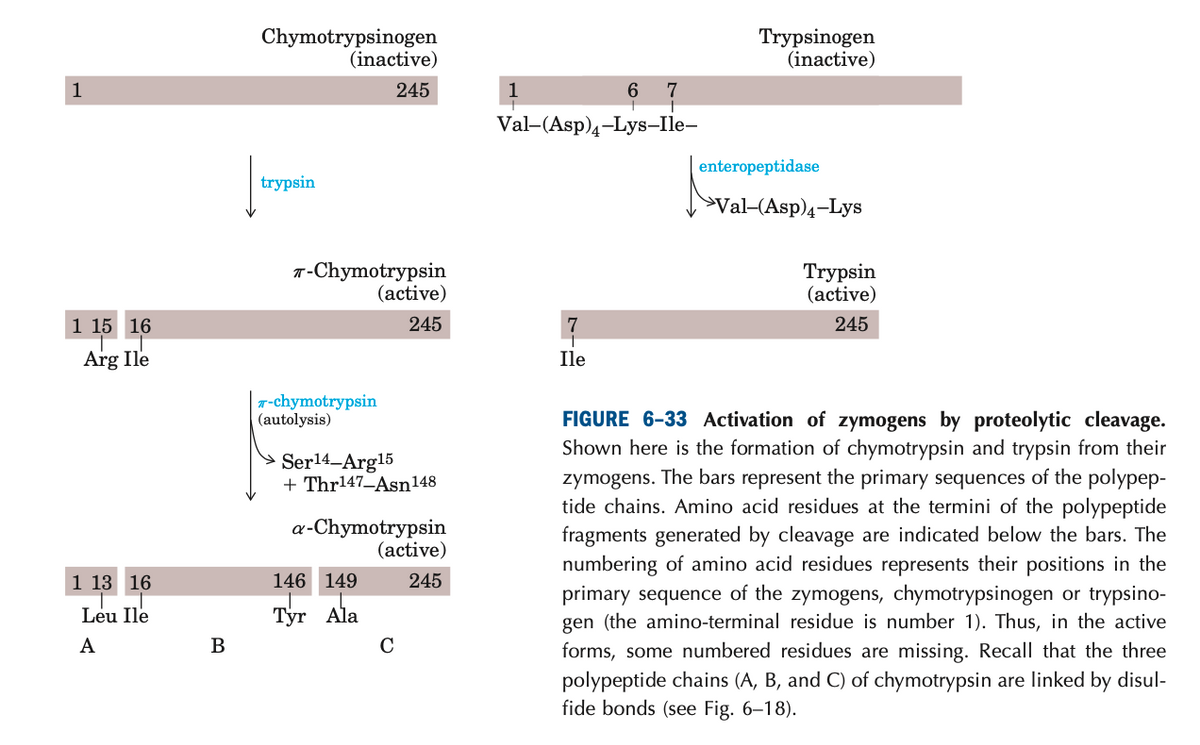
Transcribed Image Text:Chymotrypsinogen
(inactive)
Trypsinogen
(inactive)
245
1
6
Val-(Asp)4-Lys-Ile-
enteropeptidase
trypsin
>Val-(Asp)4-Lys
T-Chymotrypsin
(active)
Trypsin
(active)
1 15 16
245
7
245
Arg Ile
Ile
T-chymotrypsin
(autolysis)
FIGURE 6-33 Activation of zymogens by proteolytic cleavage.
Shown here is the formation of chymotrypsin and trypsin from their
zymogens. The bars represent the primary sequences of the polypep-
tide chains. Amino acid residues at the termini of the polypeptide
fragments generated by cleavage are indicated below the bars. The
numbering of amino acid residues represents their positions in the
Ser14-Arg15
+ Thr147_Asn148
a-Chymotrypsin
(active)
1 13 16
146 149
245
primary sequence of the zymogens, chymotrypsinogen or trypsino-
gen (the amino-terminal residue is number 1). Thus, in the active
Leu Ile
Тyr Ala
A
C
forms, some numbered residues are missing. Recall that the three
polypeptide chains (A, B, and C) of chymotrypsin are linked by disul-
fide bonds (see Fig. 6–18).
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 2 steps

Recommended textbooks for you

Human Physiology: From Cells to Systems (MindTap …
Biology
ISBN:
9781285866932
Author:
Lauralee Sherwood
Publisher:
Cengage Learning

Human Physiology: From Cells to Systems (MindTap …
Biology
ISBN:
9781285866932
Author:
Lauralee Sherwood
Publisher:
Cengage Learning