Part F - Identifying conserved residues from the BLAST alignment If an amino acid is found at the exact same location in the alignment of two homologous proteins, that amino acid is said to be "absolutely conserved." An amino acid type can also be conserved at a given position. Using the BLAST sequence alignment above, sort aligned residues into the appropriate categories. Drag the appropriate alignments to their respective bins. ► View Available Hint(s) I→L Q→E V→Q D→S Absolutely conserved W F T→M D→N Conserved by type N→G K→K A→E Not conserved Reset Help
Part F - Identifying conserved residues from the BLAST alignment If an amino acid is found at the exact same location in the alignment of two homologous proteins, that amino acid is said to be "absolutely conserved." An amino acid type can also be conserved at a given position. Using the BLAST sequence alignment above, sort aligned residues into the appropriate categories. Drag the appropriate alignments to their respective bins. ► View Available Hint(s) I→L Q→E V→Q D→S Absolutely conserved W F T→M D→N Conserved by type N→G K→K A→E Not conserved Reset Help
Biochemistry
6th Edition
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Reginald H. Garrett, Charles M. Grisham
Chapter31: Completing The Protein Life Cycle: Folding, Processing, And Degradation
Section: Chapter Questions
Problem 20P
Related questions
Question

Transcribed Image Text:The full amino acid sequence of FABP6 is
MAFTGKFEMECEKNYDEFMKLLGISSDVIEKARNFKIVTEVQQDGQDF
TWSQHYSGGHTMTNKFTVGKECNIQTMGGKTFKATVQMEGGKLVVN
FPNYHQTSEIVGDKLVEVSTIGGVTYERVSKRLA
For the next step in your analysis, you ran a BLAST search of the PDB of this sequence and found a structure that is 59% identical to the
sequence of your protein. The following is the alignment of two sequences (with your protein in blue (FABP6) and the homologous structure in
red (3ELX)):
Expect score = : 4 × 10−4² and Identities = 69/116(50%), Positives = 88/116(75%)
3ELX: 5 AFNGKWETECQEGYEPFCKLIGIPDDVIAKGRDFKLVTEIVQNGDDFTWTQYYPNNHVMT 64
AF GK + E EC++ Y+ F KL GI DVI K R+FK+VTE+ Q+G DFTW+Q+Y
FABP6: 2 AFTGKFEMECEKNYDEFMKLLGISSDVIEKARNFKIVTEVQQDGQDFTWSQHYSGGHTMT 61
3ELX: 65 NKF IVGKECDMETVGGKKFKG IVSMEGGKLT IS FPKYQQTTEISGGKLVETSTASG 120
NKF VGKEC+++T+GGK FK V MEGGKL ++FP Y QT+EI G KLVE ST G
FABP6: 62 NKFTVGKECNIQTMGGKTFKATVQMEGGKLVVNFPNYHQTSEIVGDKLVEVSTIGG
H MT
117
The identical amino acids are shown in black between the two sequences. A notation of "+" indicates a similar, but not the same, amino acid.
The sequences are arranged to show overlap and gaps in the primary amino acid sequence.
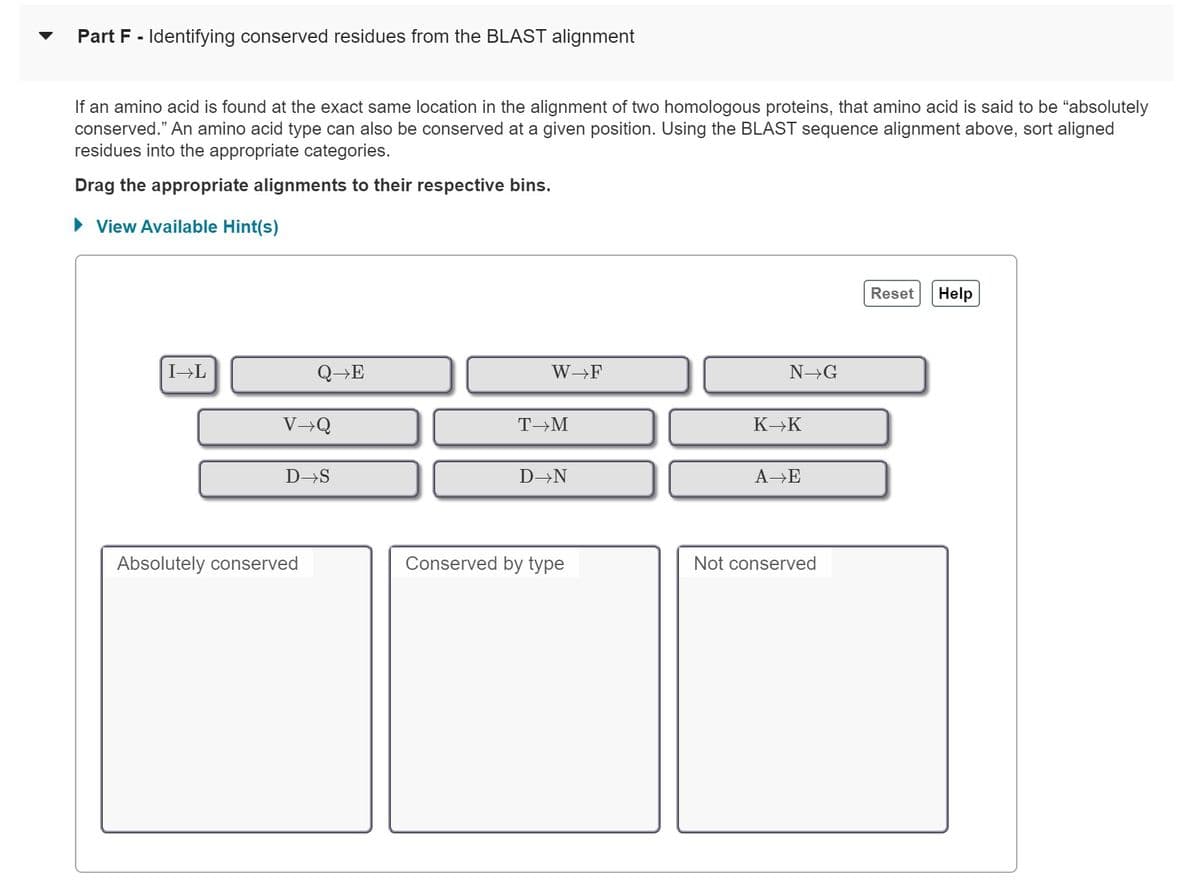
Transcribed Image Text:Part F - Identifying conserved residues from the BLAST alignment
If an amino acid is found at the exact same location in the alignment of two homologous proteins, that amino acid is said to be "absolutely
conserved." An amino acid type can also be conserved at a given position. Using the BLAST sequence alignment above, sort aligned
residues into the appropriate categories.
Drag the appropriate alignments to their respective bins.
► View Available Hint(s)
I→L
QE
V→Q
D→S
Absolutely conserved
W→F
T→M
D→N
Conserved by type
N→G
K→K
A→E
Not conserved
Reset Help
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 4 steps with 17 images

Recommended textbooks for you

Biochemistry
Biochemistry
ISBN:
9781305577206
Author:
Reginald H. Garrett, Charles M. Grisham
Publisher:
Cengage Learning

Biochemistry
Biochemistry
ISBN:
9781305577206
Author:
Reginald H. Garrett, Charles M. Grisham
Publisher:
Cengage Learning