Which phenomenon is not readily explained by the induced-fit' hypothesis, that is readily explained by the fluctuation-fit' hypothesis? O Catalysis by optimal interaction with the transition state of substrates. O Specificity of enzymes for substrates. O Lower binding affinity for products, enabling their release once chemistry has happened. O Cooperativity of binding to the remaining 3 sites of hemoglobin when oxygen binds to the first of the four sites.
Which phenomenon is not readily explained by the induced-fit' hypothesis, that is readily explained by the fluctuation-fit' hypothesis? O Catalysis by optimal interaction with the transition state of substrates. O Specificity of enzymes for substrates. O Lower binding affinity for products, enabling their release once chemistry has happened. O Cooperativity of binding to the remaining 3 sites of hemoglobin when oxygen binds to the first of the four sites.
Biochemistry
9th Edition
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Chapter1: Biochemistry: An Evolving Science
Section: Chapter Questions
Problem 1P
Related questions
Question

Transcribed Image Text:Which phenomenon is not readily explained by the 'induced-fit' hypothesis, that is readily explained by
the 'fluctuation-fit hypothesis?
Catalysis by optimal interaction with the transition state of substrates.
O Specificity of enzymes for substrates.
O Lower binding affinity for products, enabling their release once chemistry has happened.
O Cooperativity of binding to the remaining 3 sites of hemoglobin when oxygen binds to the first of the four sites.
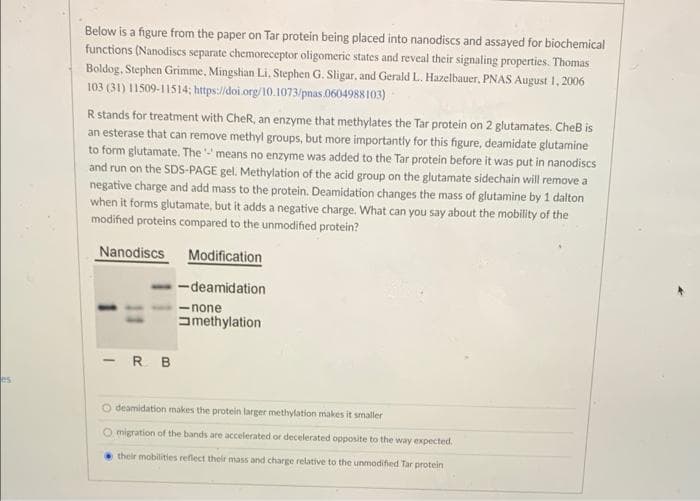
Transcribed Image Text:Below is a figure from the paper on Tar protein being placed into nanodiscs and assayed for biochemical
functions (Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Thomas
Boldog. Stephen Grimme, Mingshian Li, Stephen G. Sligar, and Gerald L. Hazelbauer, PNAS August 1, 2006
103 (31) 11509-11514; https://doi.org/10.1073/pnas.0604988 103)-
R stands for treatment with CheR, an enzyme that methylates the Tar protein on 2 glutamates. CheB is
an esterase that can remove methyl groups, but more importantly for this figure, deamidate glutamine
to form glutamate. The means no enzyme was added to the Tar protein before it was put in nanodiscs
and run on the SDS-PAGE gel. Methylation of the acid group on the glutamate sidechain will remove a
negative charge and add mass to the protein. Deamidation changes the mass of glutamine by 1 dalton
when it forms glutamate, but it adds a negative charge. What can you say about the mobility of the
modified proteins compared to the unmodified protein?
Nanodiscs
Modification
-deamidation
- none
amethylation
-R B
O deamidation makes the protein larger methylation makes it smaller
O migration of the bands are accelerated or decelerated opposite to the way expected.
their mobilities reflect their mass and charge relative to the unmodified Tar protein
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution!
Trending now
This is a popular solution!
Step by step
Solved in 2 steps

Recommended textbooks for you

Biochemistry
Biochemistry
ISBN:
9781319114671
Author:
Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:
W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:
9781464126116
Author:
David L. Nelson, Michael M. Cox
Publisher:
W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul…
Biochemistry
ISBN:
9781118918401
Author:
Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:
WILEY

Biochemistry
Biochemistry
ISBN:
9781319114671
Author:
Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:
W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:
9781464126116
Author:
David L. Nelson, Michael M. Cox
Publisher:
W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul…
Biochemistry
ISBN:
9781118918401
Author:
Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:
WILEY

Biochemistry
Biochemistry
ISBN:
9781305961135
Author:
Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:
Cengage Learning

Biochemistry
Biochemistry
ISBN:
9781305577206
Author:
Reginald H. Garrett, Charles M. Grisham
Publisher:
Cengage Learning

Fundamentals of General, Organic, and Biological …
Biochemistry
ISBN:
9780134015187
Author:
John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:
PEARSON