
Biochemistry: Concepts and Connections
1st Edition
ISBN: 9780321839923
Author: Dean R. Appling, Spencer J. Anthony-Cahill, Christopher K. Mathews
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 8, Problem 11P
The following data describe the catalysis of cleavage of peptide bonds small peptides by the enzyme elastase.
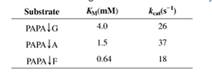
The arrow indicates the peptide bond cleaved each case.
a. If a mixture of these three substrates was presented to elastase with the concentration of each peptide equal to 0.5 mM, which would be digested most rapidly? Which most slowly? (Assume enzyme present in excess.)
b. On basis of these data, suggest what features of amino acid sequence dictate the specificity of proteolytic cleavage by elastase.
c. Elastase is closely related to chymotrypsin. Suggest two kinds of amino acid residues you might expect to find in or near the active site.
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
The following data describe the catalysis of cleavage of peptide bonds in
small peptides by the enzyme elastase.
Substrate
Ka(mM) ka(s")
PAPAIG
4.0
26
PAPA JA
1.5
37
PAPA ĮF
0.64
18
The arrow indicates the peptide bond cleaved in each case.
(a) If a mixture of these three substrates was presented to elastase with the
concentration of each peptide equal to 0.5 mm, which would be digested
most rapidly? Which most slowly? (Assume enzyme is present in excess.)
(b) On the basis of these data, suggest what features of amino acid sequence
dictate the specificity of proteolytic cleavage by elastase.
(c) Elastase is closely related to chymotrypsin. Suggest two kinds of amino
acid residues you might expect to find in or near the active site.
The following steps were performed using enzyme cleavage of a peptide to determine its amino acid sequence.
Step 1. FDNB yield DNB-Gly
Step 2. Treatment with trypsin yield 3 fragments: Tyr-Leu-Asp-Arg; Gly-Ser-Ala-Lys; Trp-Gly-Ser-Met
Step 3. Treatment with pepsin gave the same 3 peptide fragments.
What is the sequence of the peptide?
The trypsin enzyme is able to hydrolyze a peptide substate at the carboxyl
side of an Arg or Lys residue. However, such a reaction can be also
influenced by the amino acid residue that follows Arg or Lys. Given the
Michaelis-Menton plots obtained for two substrates (substrate A: Ser-Val-
Arg-Pro; substrate B: Ser-Val-Arg-Phe), the Km of the enzyme is higher
for:
20
Substrate A
Assume
that these
curves do
not reach
the same
limit.
15
Substrate E
0.001
0.002
0.003
(Substrate) (molar)
Inconclusive
Substrate B
Both Substrate A and B
Intitial velocity
(micromoles/literisecond)
Chapter 8 Solutions
Biochemistry: Concepts and Connections
Ch. 8 - Prob. 1PCh. 8 - The enzyme urease catalyzes the hydrolysis of urea...Ch. 8 - An enzyme contains an active site aspartic acid...Ch. 8 - The folding and unfolding rate constants for a...Ch. 8 - In some reactions, in which a protein molecule is...Ch. 8 - Would you expect an “enzyme” designed to bind to...Ch. 8 - The initial rate for an enzyme-catalyzed reaction...Ch. 8 - a. If the total enzyme concentration in Problem 7...Ch. 8 - Prob. 9PCh. 8 - Prob. 10P
Ch. 8 - The following data describe the catalysis of...Ch. 8 - At 37 oC, the serine protease subtilisin has kcat...Ch. 8 - The accompanying figure shows three...Ch. 8 - The steady-state kinetics of an enzyme are studied...Ch. 8 - The same enzyme as in Problem 14 is studied in the...Ch. 8 - Enalapril is an anti-hypertension “pro-drug"...Ch. 8 - Initial rate data for an enzyme that obeys...Ch. 8 - Prob. 18PCh. 8 - Suggest the effects of each of the following...Ch. 8 - The inhibitory effect of an uncompetitive...Ch. 8 - Prob. 21P
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biochemistry and related others by exploring similar questions and additional content below.Similar questions
- The tetrapeptide Cys-Trp-Lys-Pro was digested with chymotrypsin and adjusted to pH=0.5 to fully protonate all digestion products. The digestion products were then applied to a strong cation exchange column (sulfonic acid). The products were eluted from the column using a pH gradient from 0.5 to 13. Write the primary amino acid sequence of the products in the order they would elute from the column. Indicate the pH value at which each product elutes from the columnarrow_forwardA peptide with the primary structure Lys-Arg-Pro-Leu-Ile-Asp-Gly-Ala must be synthesized by the methods developed by Merrifield. Calculate the percentage of the peptides synthesized that will be full length and have the correct sequence if the addition of each amino acid residue is 96% efficient. Do the calculation a second time but assume a 99% efficiency for each cycle. full-length peptides with the correct sequence if 96% efficient: full-length peptides with the correct sequence if 99% efficient: % %arrow_forwardDetermine the sequence of the following 10-mer of ferritin using the data below. i. Complete acid hydrolysis of the 10-mer results in identification of 1A, 1D, 1E, 1F, 1H, 1K, 2L, 1T, 1Y ii. Digestion of the 10-mer with chymotrypsin results in 3 fragments containing (D, H, K, T), (F, L) and (A, E, L, Y) iii. Digestion of the 10-mer with V8 protease, which cuts on the C-side of acidic amino acids, results in three fragments containing (D, F, L, Y), (A, E, L) and (H, K, T) iv. Digestion with trypsin results in a fragment H-T and a fragment with all the other amino acids v. Treatment of the 10-mer with FDNB followed by complete acid hydrolysis results in the following two derivatives, DNP-1 and DNP-2 vi. Treatment of the 10-mer with a carboxypeptidase results in a free threoninearrow_forward
- a. An oligopeptide ALVGALGATPTPQMWSHSWRGVSIKS was digested with trypsin.Which method would be most appropriate for separating the products: ion exchange or gel filtration chromatography? Explain.b. Suppose that the peptide was digested with cyanogen bromide. What would be the optimal separation technique? Explainarrow_forwardThe OXA-M290 protein is next purified by size exclusion chromatography. To determine the best type of size exclusion resin to use, the size of OXA-M290 must first be determined. Earlier, you determined the amino acid sequence of OXA-M290 (MRVLALSAVFLVASIIGMPAVAKEWQENKSWNAHFTEHKSQGVVVLWNENKQQGFTNNLKRANQAFLPASSAKIPNSLIALDLGVVKDEHQVFKWDGQTRDIATWNRDHNLITAMKYSVVPVYQEFARQIGEARMSKMLHAFDYGNEDISGNVDSFWLDGGIRISATEQISFLRKLYHNKLHVSERSQRIVKQAMLTEANGDYIIRAKTGYDTKIGWWVGWVELDDNVWFFAMNMDMPTSDGLGLRQAITKEVLKQEKIIP). Based on the amino acid sequence, what is the molecular weight of this protein? You can use the free ProtParam tool (https://web.expasy.org/protparam/) to calculate the molecular weights of proteins. Make sure to include units in your answer. Note: The amino acid sequence reported earlier does not include the His-tag that was added to OXA-M290 by the pET-28a vector. However, you do not need to consider the amino acids in the His-tag in your answer to this question. For Context ONLY: For…arrow_forwardSolve (a) , (b), (c), and (d). All is one problem.arrow_forward
- Based on the following image, what is the N-terminal AA residue, C-terminal AA residue? What is also the sequence of peptide 2 and peptide 3? Also identify the overall amino acid sequencearrow_forwardCarboxypeptidase Y (CPY) contains ten cysteine residues that form five disulfide bonds in the native structure of CPY. Suppose CPY is reduced and unfolded in urea.Part AIf the reduced unfolded protein were oxidized prior to the removal of the urea, what fraction of the resulting mixture would you expect to possess native disulfide bonds?Express your answer using three significant figures.arrow_forwardConsider the peptides Pro-Gin-Val-Phe-His-Asp-Cys and His-Gln-Pro-Cys-Asp-Phe-Val. How do these two peptides differ? (Select all that apply.) The two peptides have different compositions. The two peptides have different isoelectric points. The two peptides have different titration curves. The two peptides differ in amino acid sequence. [References] If you were to have a mythical amino acid based on glutamic acid, but one in which the hydrogen that is attached to the y-carbon were replaced by another amino group, what would be the predominant form of this amino acid at pH 12 if the pK, value were 10 for the unique amino group? (Select all that apply.) Both of the carboxyl groups are deprotonated. The amino acid-carries a negative 2 charge. The amino acid carries a negative 4 charge. The amino groups are in the form -NH". Both of the amino groups are deprotonated.arrow_forward
- Determine the pI of the peptide H2N-Ala-Lys-Ser-Arg-COOH at pH 11, please explain why some pKas are used in the solution of the problem while others are not.arrow_forwardAmino acid alaninearrow_forwardThe globular proteins tend to have a high-dimensional folding in their higher-order structure g providing very limited analysis when conducting peptide analysis. Explain what steps would be suitable to prepare a successful chromatographic peptide mapping?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning
Biomolecules - Protein - Amino acids; Author: Tutorials Point (India) Ltd.;https://www.youtube.com/watch?v=ySNVPDHJ0ek;License: Standard YouTube License, CC-BY